Epigenomic Data Analysis 2017 Module 3 Lab
Module 3: Introduction to WGBS and Analysis
by Guillaume Bourque, PhD
Important notes:
- Please refer to the following guide for instructions on how to connect to the Compute Canada cluster: Compute Canada instructions
- The user lect99 below is provided as an example. You should replace it by the username that was assigned to you at the beginning of the workshop.
Introduction
Description of the lab
This module will cover the basics of Bisulfite-sequencing data analysis including data visualization in IGV.
Local software that we will use
- ssh
- IGV
Tutorial
Getting started
Connect to the Compute Canada cluster
ssh lect99@workshop103.ccs.usherbrooke.ca
You will be in your home folder. At this step, before continuing, please make sure that you followed the instructions in the section “The first time you log in” of the Compute Canada instructions. If you don’t, compute jobs will not execute normally.
Prepare directory for module 3
rm -rf ~/module3
mkdir -p ~/module3
cd ~/module3
Copy data for module 3
mkdir data
cp /home/partage/epigenomics/wgb-seq/data/* data/.
Check the files
By typing ls data you should see something similar to this
[lect99@workshop103 module3]$ ls data
iPSC_1.1.fastq iPSC_1.2.fastq iPSC_2.1.fastq iPSC_2.2.fastq
What do the “.1” and “.2” in the file names mean?
Map using bismark
We will now process and map the reads using Bismark.
module load mugqic/bismark/0.16.1
bismark --bowtie2 -n 1 /home/partage/epigenomics/wgb-seq/genome/ -1 data/iPSC_1.1.fastq -2 data/iPSC_1.2.fastq
The --bowtie2 is to use the mapping algorithm bowtie.
The -n 1 defines the maximum number of mismatches permitted in the seed.
The /home/partage/epigenomics/wgb-seq/genome/ specifies the reference genome to use.
The -1 data/iPSC_1.1.fastq specifies the location of read 1. Idem for -2 which specifies read 2.
For more details, please refer to the Bismark user guide.
Check status of your job
Bismark first loads the reference genome one chromosome at a time. You should be able to follow progress.
Check output messages
Is this what you expected?
Map (again) using bismark
rm iPSC_*
module load mugqic/bismark/0.16.1 ; module load mugqic/bowtie2/2.2.4 ; module load mugqic/samtools/1.3
bismark --bowtie2 -n 1 /home/partage/epigenomics/wgb-seq/genome/ -1 data/iPSC_1.1.fastq -2 data/iPSC_1.2.fastq
Check files
At the end, you should have something similar to
[lect99@workshop103 module3]$ ls -ltr
total 13662
drwxr-xr-x 2 lect03 lect 6 Jun 15 07:12 data
-rw-r--r-- 1 lect03 lect 13964434 Jun 15 07:36 iPSC_1.1_bismark_bt2_pe.bam
-rw-r--r-- 1 lect03 lect 1839 Jun 15 07:36 iPSC_1.1_bismark_bt2_PE_report.txt
Let’s look at the report
less iPSC_1.1_bismark_bt2_PE_report.txt
Prepare files for loading in IGV
We need to sort the bam file and prepare an index so we will be able to load in IGV. We will use the program samtools for this.
module load mugqic/samtools/1.3
samtools sort iPSC_1.1_bismark_bt2_pe.bam -o iPSC_1.1_bismark_bt2_pe_sorted.bam
samtools index iPSC_1.1_bismark_bt2_pe_sorted.bam
Check files
At the end, you should have something similar to
[lect99@workshop103 module3]$ ls -ltr
total 25471
drwxr-xr-x 2 lect03 lect 6 Jun 15 07:12 data
-rw-r--r-- 1 lect03 lect 13964434 Jun 15 07:36 iPSC_1.1_bismark_bt2_pe.bam
-rw-r--r-- 1 lect03 lect 1839 Jun 15 07:36 iPSC_1.1_bismark_bt2_PE_report.txt
-rw-r--r-- 1 lect03 lect 11653598 Jun 15 07:39 iPSC_1.1_bismark_bt2_pe_sorted.bam
-rw-r--r-- 1 lect03 lect 1967480 Jun 15 07:39 iPSC_1.1_bismark_bt2_pe_sorted.bam.bai
Copy files to your local computer to view in IGV
Using a different terminal window that is not connected to the server (if you are using Mac/Linux) or WinSCP (if you are using Windows), retrieve the iPSC_1.1_bismark_bt2_pe_sorted.bam and iPSC_1.1_bismark_bt2_pe_sorted.bam.bai
scp lect%%@workshop103.ccs.usherbrooke.ca:/home/lect%%/module3/iPSC_1.1_bismark_bt2_pe_sorted.bam* .
Where you need to replace the two places with “%%” by your student number.
Load data and explore using IGV
Launch IGV on your computer.
If you haven’t installed it yet, please get it here IGV download.
Load your sorted bam and index file in IGV using File -> Load from file.
Go to:
chr3:43,375,889-45,912,052
And zoom in until you see something.
For instance go to:
chr3:44,513,532-44,523,018
You should see something like
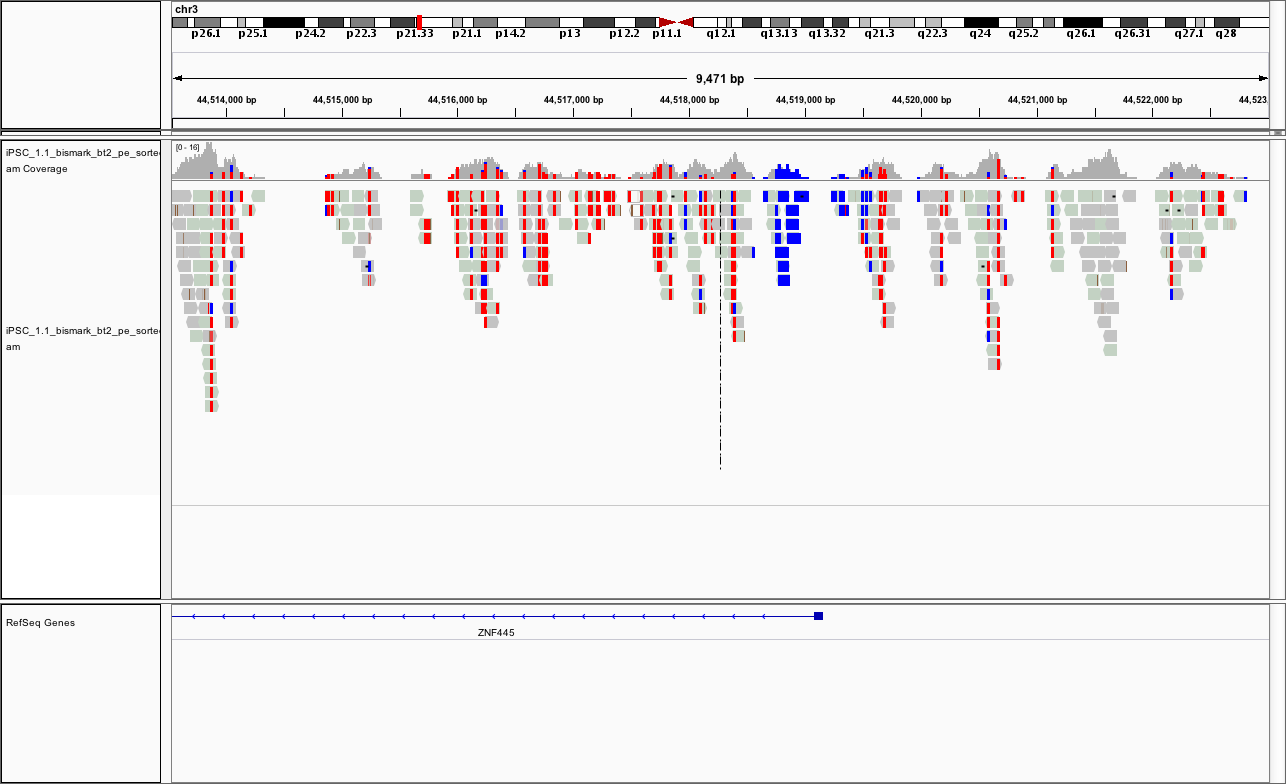
If it looks different, can you change the way the colors are displayed?
Repeat for the other replicate
Map using bismark
module load mugqic/bismark/0.16.1 ; module load mugqic/bowtie2/2.2.4 ; module load mugqic/samtools/1.3
bismark --bowtie2 -n 1 /home/partage/epigenomics/wgb-seq/genome/ -1 data/iPSC_2.1.fastq -2 data/iPSC_2.2.fastq
Did you need to repeat the module load commands?
And what context would you need to repeat them?
Prepare files for IGV
samtools sort iPSC_2.1_bismark_bt2_pe.bam -o iPSC_2.1_bismark_bt2_pe_sorted.bam
samtools index iPSC_2.1_bismark_bt2_pe_sorted.bam
Check files
At this point you should have something like
[lect99@workshop103 module3]$ ls -ltr
total 56553
drwxr-xr-x 2 lect03 lect 6 Jun 15 07:12 data
-rw-r--r-- 1 lect03 lect 13964434 Jun 15 07:36 iPSC_1.1_bismark_bt2_pe.bam
-rw-r--r-- 1 lect03 lect 1839 Jun 15 07:36 iPSC_1.1_bismark_bt2_PE_report.txt
-rw-r--r-- 1 lect03 lect 11653598 Jun 15 07:39 iPSC_1.1_bismark_bt2_pe_sorted.bam
-rw-r--r-- 1 lect03 lect 1967480 Jun 15 07:39 iPSC_1.1_bismark_bt2_pe_sorted.bam.bai
-rw-r--r-- 1 lect03 lect 17226630 Jun 15 07:52 iPSC_2.1_bismark_bt2_pe.bam
-rw-r--r-- 1 lect03 lect 1839 Jun 15 07:52 iPSC_2.1_bismark_bt2_PE_report.txt
-rw-r--r-- 1 lect03 lect 14046458 Jun 15 07:53 iPSC_2.1_bismark_bt2_pe_sorted.bam
-rw-r--r-- 1 lect03 lect 2064608 Jun 15 07:53 iPSC_2.1_bismark_bt2_pe_sorted.bam.bai
Generate methylation profiles from the bam files
So far we have only mapped the reads using bismark. We can now generate methylation profiles using the following command
bismark_methylation_extractor --bedGraph iPSC_1.1_bismark_bt2_pe.bam
Do the same for the other replicate
bismark_methylation_extractor --bedGraph iPSC_2.1_bismark_bt2_pe.bam
Check files
At this point you should have something like
[lect03@workshop103 module3]$ ls -ltr
total 103111
drwxr-xr-x 2 lect03 lect 6 Jun 15 07:12 data
-rw-r--r-- 1 lect03 lect 13964434 Jun 15 07:36 iPSC_1.1_bismark_bt2_pe.bam
-rw-r--r-- 1 lect03 lect 1839 Jun 15 07:36 iPSC_1.1_bismark_bt2_PE_report.txt
-rw-r--r-- 1 lect03 lect 11653598 Jun 15 07:39 iPSC_1.1_bismark_bt2_pe_sorted.bam
-rw-r--r-- 1 lect03 lect 1967480 Jun 15 07:39 iPSC_1.1_bismark_bt2_pe_sorted.bam.bai
-rw-r--r-- 1 lect03 lect 17226630 Jun 15 07:52 iPSC_2.1_bismark_bt2_pe.bam
-rw-r--r-- 1 lect03 lect 1839 Jun 15 07:52 iPSC_2.1_bismark_bt2_PE_report.txt
-rw-r--r-- 1 lect03 lect 14046458 Jun 15 07:53 iPSC_2.1_bismark_bt2_pe_sorted.bam
-rw-r--r-- 1 lect03 lect 2064608 Jun 15 07:53 iPSC_2.1_bismark_bt2_pe_sorted.bam.bai
-rw-r--r-- 1 lect03 lect 3317542 Jun 15 07:54 CpG_OT_iPSC_1.1_bismark_bt2_pe.txt
-rw-r--r-- 1 lect03 lect 3233404 Jun 15 07:54 CpG_OB_iPSC_1.1_bismark_bt2_pe.txt
-rw-r--r-- 1 lect03 lect 47352591 Jun 15 07:54 CHH_OT_iPSC_1.1_bismark_bt2_pe.txt
-rw-r--r-- 1 lect03 lect 45655164 Jun 15 07:54 CHH_OB_iPSC_1.1_bismark_bt2_pe.txt
-rw-r--r-- 1 lect03 lect 15632135 Jun 15 07:54 CHG_OT_iPSC_1.1_bismark_bt2_pe.txt
-rw-r--r-- 1 lect03 lect 14933671 Jun 15 07:54 CHG_OB_iPSC_1.1_bismark_bt2_pe.txt
-rw-r--r-- 1 lect03 lect 827 Jun 15 07:54 iPSC_1.1_bismark_bt2_pe_splitting_report.txt
-rw-r--r-- 1 lect03 lect 13068 Jun 15 07:54 iPSC_1.1_bismark_bt2_pe.M-bias.txt
-rw-r--r-- 1 lect03 lect 366080 Jun 15 07:54 iPSC_1.1_bismark_bt2_pe.bismark.cov.gz
-rw-r--r-- 1 lect03 lect 388554 Jun 15 07:54 iPSC_1.1_bismark_bt2_pe.bedGraph.gz
-rw-r--r-- 1 lect03 lect 2912404 Jun 15 07:57 CpG_OT_iPSC_2.1_bismark_bt2_pe.txt
-rw-r--r-- 1 lect03 lect 2778922 Jun 15 07:57 CpG_OB_iPSC_2.1_bismark_bt2_pe.txt
-rw-r--r-- 1 lect03 lect 53931897 Jun 15 07:57 CHH_OT_iPSC_2.1_bismark_bt2_pe.txt
-rw-r--r-- 1 lect03 lect 52417972 Jun 15 07:57 CHH_OB_iPSC_2.1_bismark_bt2_pe.txt
-rw-r--r-- 1 lect03 lect 15629482 Jun 15 07:57 CHG_OT_iPSC_2.1_bismark_bt2_pe.txt
-rw-r--r-- 1 lect03 lect 15044910 Jun 15 07:57 CHG_OB_iPSC_2.1_bismark_bt2_pe.txt
-rw-r--r-- 1 lect03 lect 828 Jun 15 07:58 iPSC_2.1_bismark_bt2_pe_splitting_report.txt
-rw-r--r-- 1 lect03 lect 13234 Jun 15 07:58 iPSC_2.1_bismark_bt2_pe.M-bias.txt
-rw-r--r-- 1 lect03 lect 340966 Jun 15 07:58 iPSC_2.1_bismark_bt2_pe.bedGraph.gz
-rw-r--r-- 1 lect03 lect 322840 Jun 15 07:58 iPSC_2.1_bismark_bt2_pe.bismark.cov.gz
Uncompress the bedGraph files
gunzip iPSC_1.1_bismark_bt2_pe.bedGraph.gz
gunzip iPSC_2.1_bismark_bt2_pe.bedGraph.gz
Transfer the files to your local computer
Using a different terminal window that is not connected to the server (if you are using Mac/Linux) or WinSCP (if you are using Windows), retrieve the iPSC_2.1_bismark_bt2_pe_sorted.bam and iPSC_2.1_bismark_bt2_pe_sorted.bam.bai
scp lect%%@workshop103.ccs.usherbrooke.ca:/home/lect%%/module3/iPSC_2.1_bismark_bt2_pe_sorted.bam* .
Also transfer the bedGraphs
scp lect%%@workshop103.ccs.usherbrooke.ca:/home/lect%%/module3/*bedGraph* .
Where you need to replace the two places with “%%” by your student number.
Load all the data in IGV
Load iPSC_1.1_bismark_bt2_pe.bedGraph in IGV using File -> Load from file.
Load iPSC_2.1_bismark_bt2_pe_sorted.bam in IGV using File -> Load from file.
Load iPSC_2.1_bismark_bt2_pe.bedGraph in IGV using File -> Load from file.
At this point, if you load the region chr3:44,513,532-44,523,018 you should see something like
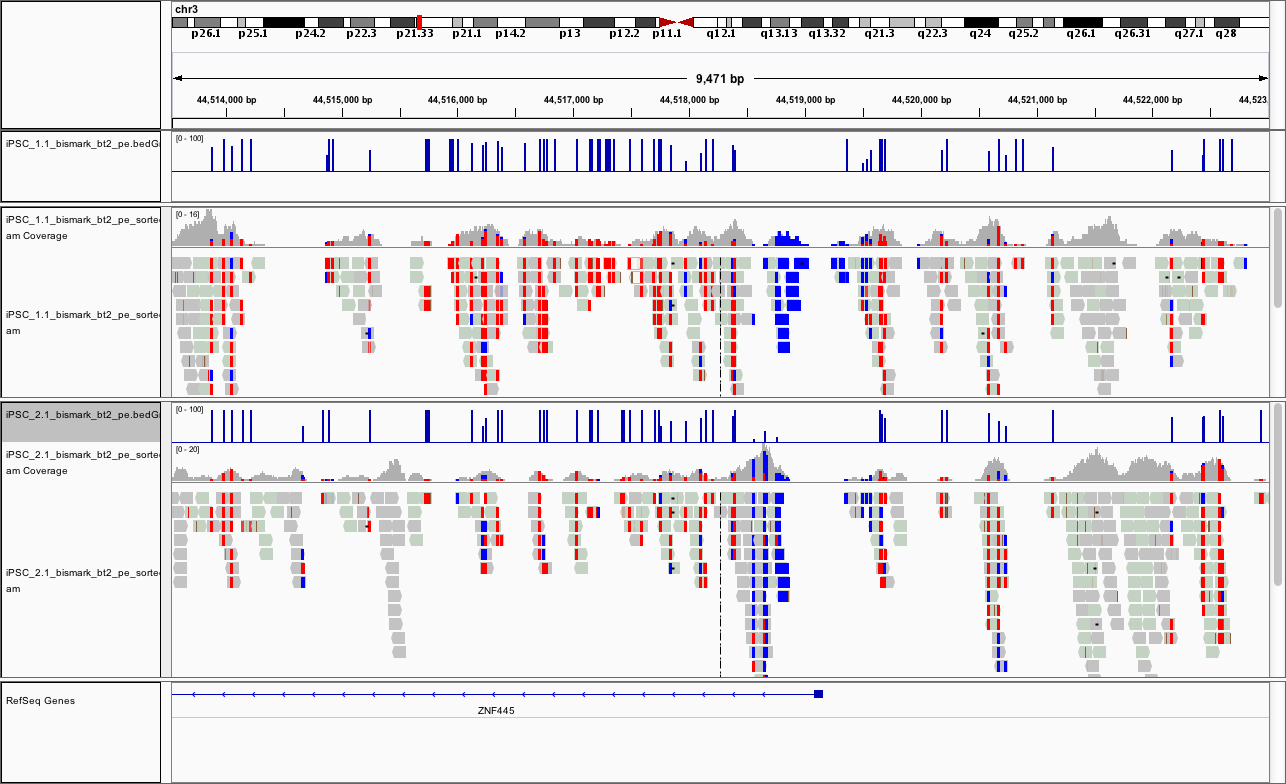
This promoter looks to be hypomethylated.
Can you find a promoter that is hypermethylated?
How about chr3:44,274,770-44,293,744?
Do you how to load CpG islands annotation?
Congrats, you’re done!
Continue exploring on your own…