Working with Big Cancer Data in the Collaboratory Cloud 2017 Module 1 Lab
This lab was created by George Mihaiescu
Introduction
Description of the lab
Welcome to the lab for Working with Big Cancer Data in the Collaboratory Cloud!
This lab will take you through the steps required to setup and configure your virtual machine with Docker packages, and will show you how to access data in the Cloud.
After this lab, you will be able to:
- Setup and launch a virtual machine
- Install Docker and launch a container inside a virtual machine
- Setup the ICGC storage client packaged as a docker container
- Learn how to access protected data in the Cloud using the ICGC storage client
Things to know before you start:
The lab may take between 1-2 hours, depending on your familiarity with Cloud Computing and Linux command line.
The project used by the lab is shared by all students, so please do not perform any actions on other students’ virtual machines or other resources.
Requirements
- Laptop connected to Internet
- Web browser
- SSH client (pre-installed on Linux or Mac, use the free Putty on Windows)
- Collaboratory credentials (to be provided by lab assistants)
Note: The Collaboratory credentials you are given for the workshop will only work during the workshop.
Log In to the Collaboratory
In your browser, go to https://console.cancercollaboratory.org. Log in using your provided credentials.

Once logged in, the first page open will be the “Overview Page” that shows the resources available to your project, as well as historical usage.

Before Launching a Virtual Machine
Create a SSH Key-pair
In the bar on the left side of the page, under Project, Compute tab, click on “Access and Security” and then on the “Create Key Pair” button. Name your key-pair and click on the “Create Key Pair” button. This will prompt you to save a file to your computer. Take note where you save this file because you will need to find it later.

On Windows with PuTTY
Openstack currently creates key-pairs that only work natively with Mac and Linux, so if you are using a Windows computer you will have to convert the pem key provided by Openstack to a format recognized by Putty, the free Windows SSH client utility. In order to do this, you need to start the PuTTY Key Generator https://www.ssh.com/ssh/putty/windows/puttygen and follow the instructions about converting the key provided at https://github.com/naturalis/openstack-docs/wiki/Howto:-Creating-and-using-OpenStack-SSH-keypairs-on-Windows, or in the screenshot below:

Customize Your Security Groups - this step is not needed because we already customized the security group with a rule allowing all SSH traffic, but it’s here for future reference
You will need to know your IP address for this. To find your IP address, open a new tab or window and go to Google and search for “what is my ip”.

Return to the Collaboratory page. Select the “Security Groups” tab and click on the “Create Security Group” button. Name your security group (ie ssh_yourname) and write a description. Click on “Create Security Group”.
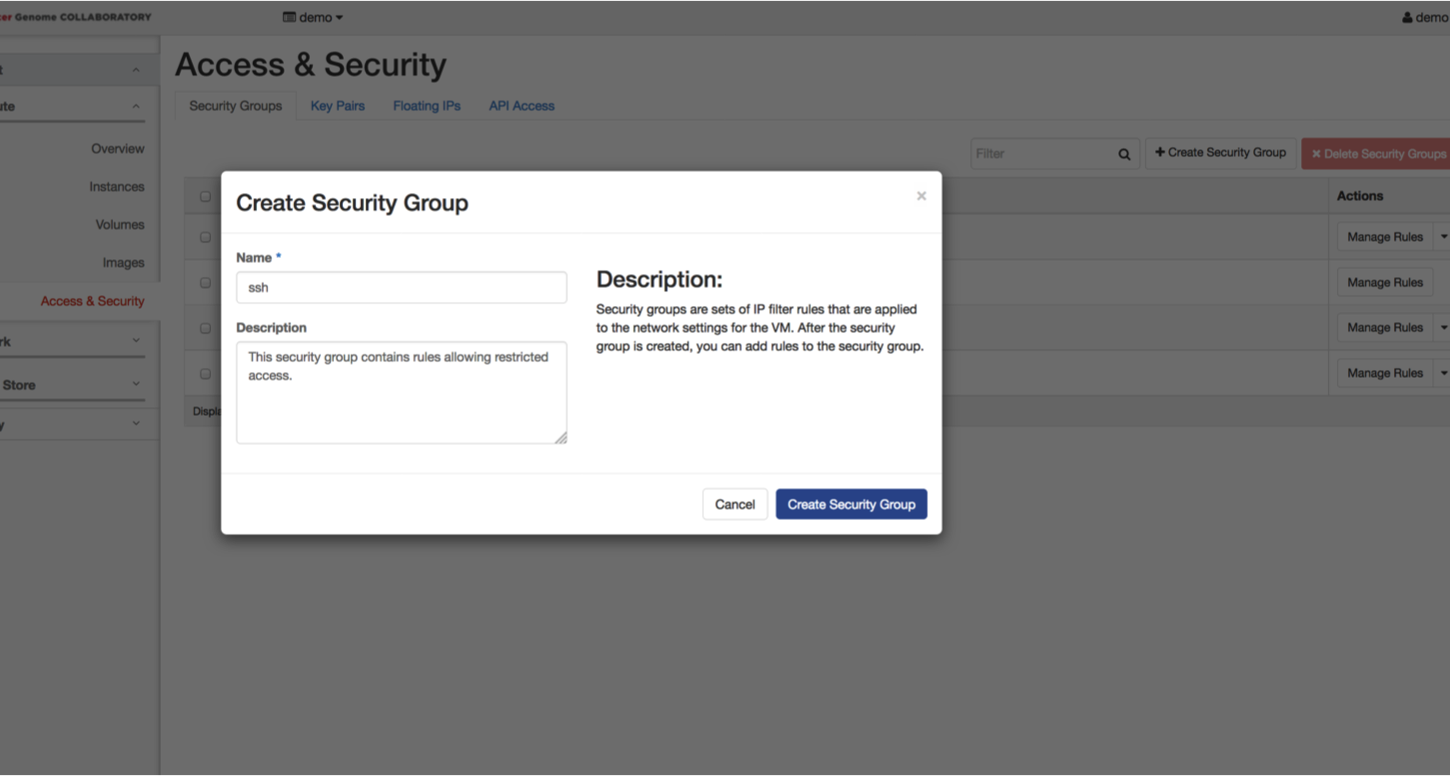
For this lab, you will need to allow SSH access from your IP address, as well as HTTP. Beside the name for the security group you just created, click on “Manage Rules”. Click on the “Add Rule” button.
In the dropdown menus and boxes, select or enter the following to allow SSH traffic:
- Custom TCP Rule
- Ingress
- Port
- 22
- CIDR
- your IP address
Repeat this step and add a second rule allowing access to TCP port 80:
- Custom TCP Rule
- Ingress
- Port
- 80
- CIDR
- your IP address

Note: If you want to allow access to the entire Internet, use “0.0.0.0/0” as the source when adding the security rules. This is insecure because it opens up your virtual machine to potential malicious traffic, so please use causiously and make sure you secure your virtual machine before doing so.
Launch your virtual machine
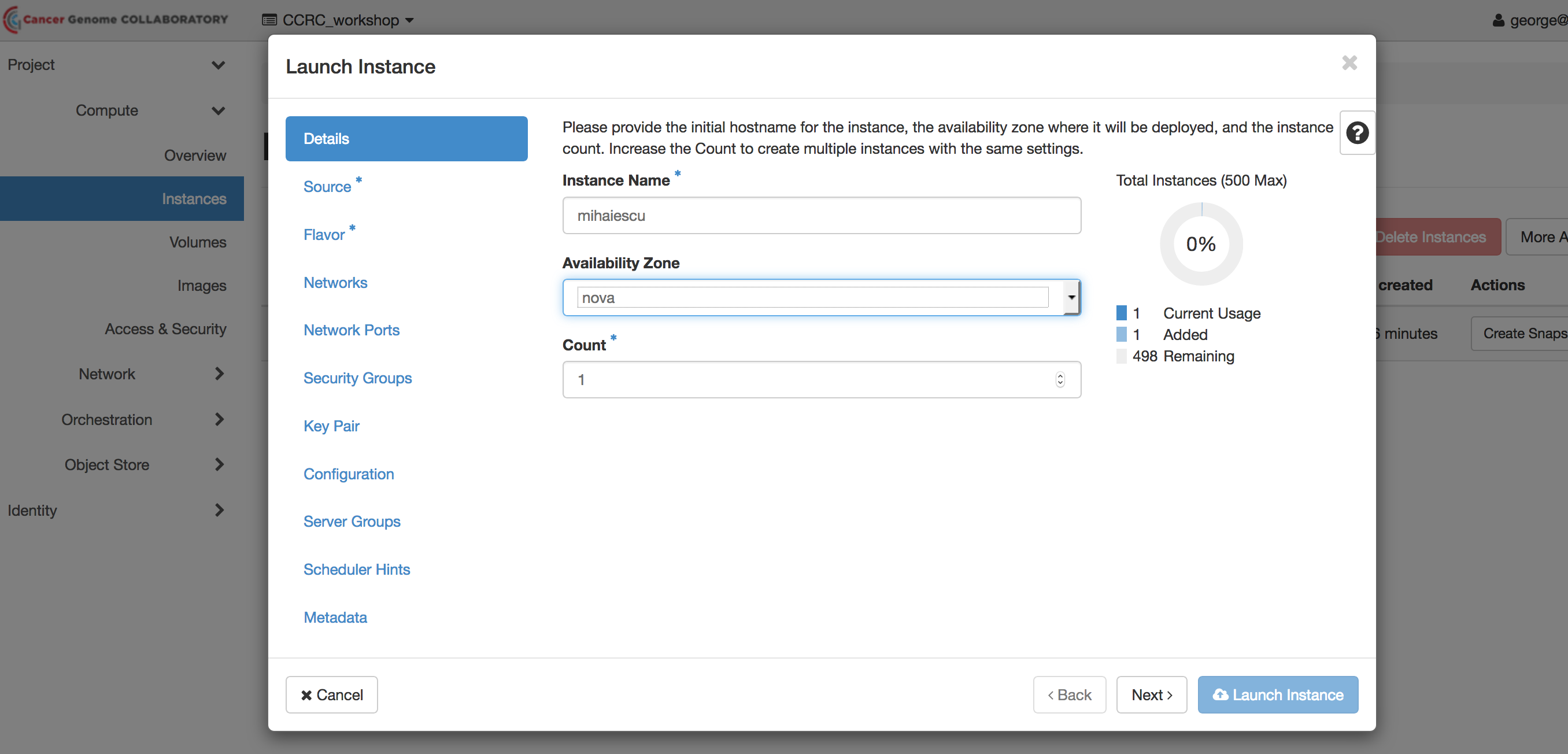
In the menu on the left, select “Instances.” Click on the “Launch Instance” button and follow the same screens as in the previous lab to start an instance with the following settings:
** Instance Name: use your last name **
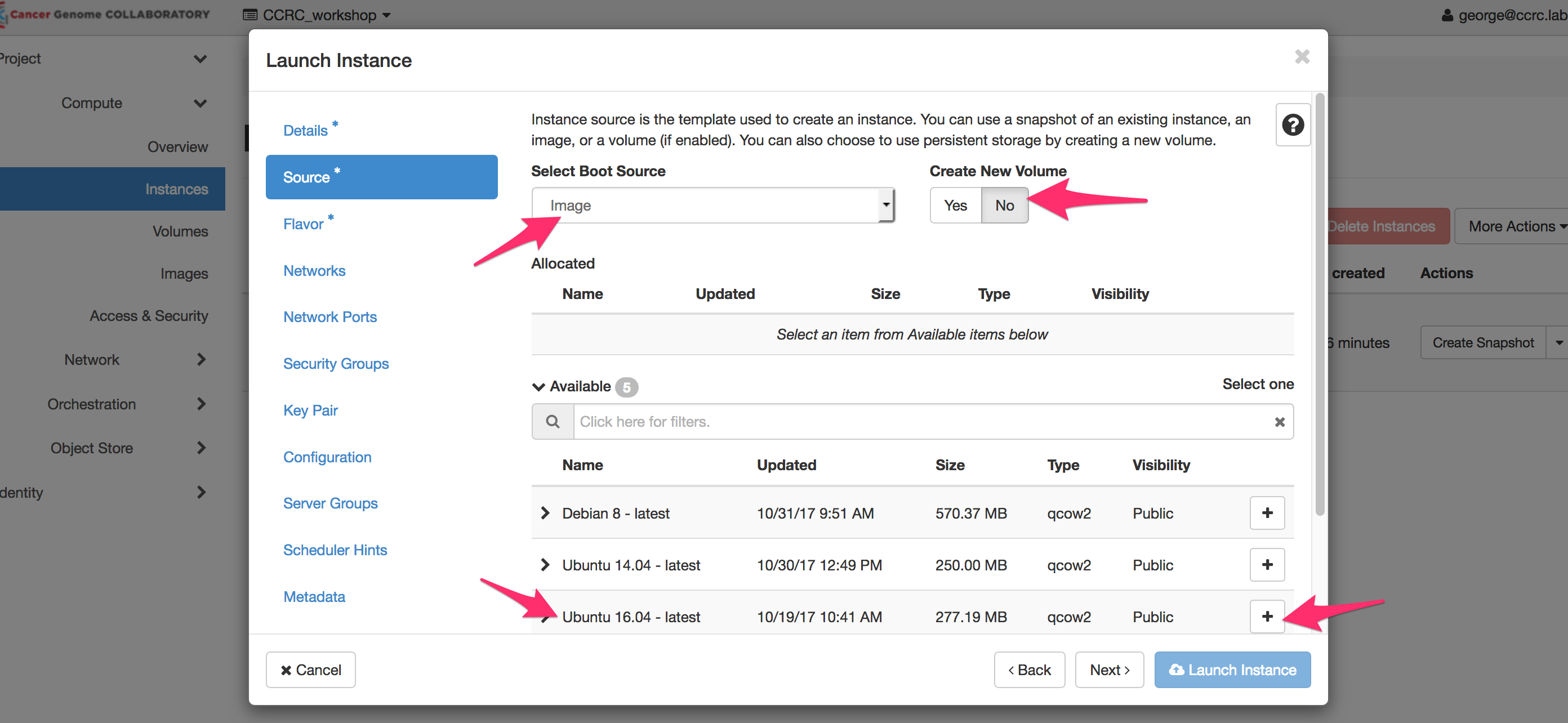
** Select Boot Source: Image, Create New Volume: No, Ubuntu 16.04 - latest**
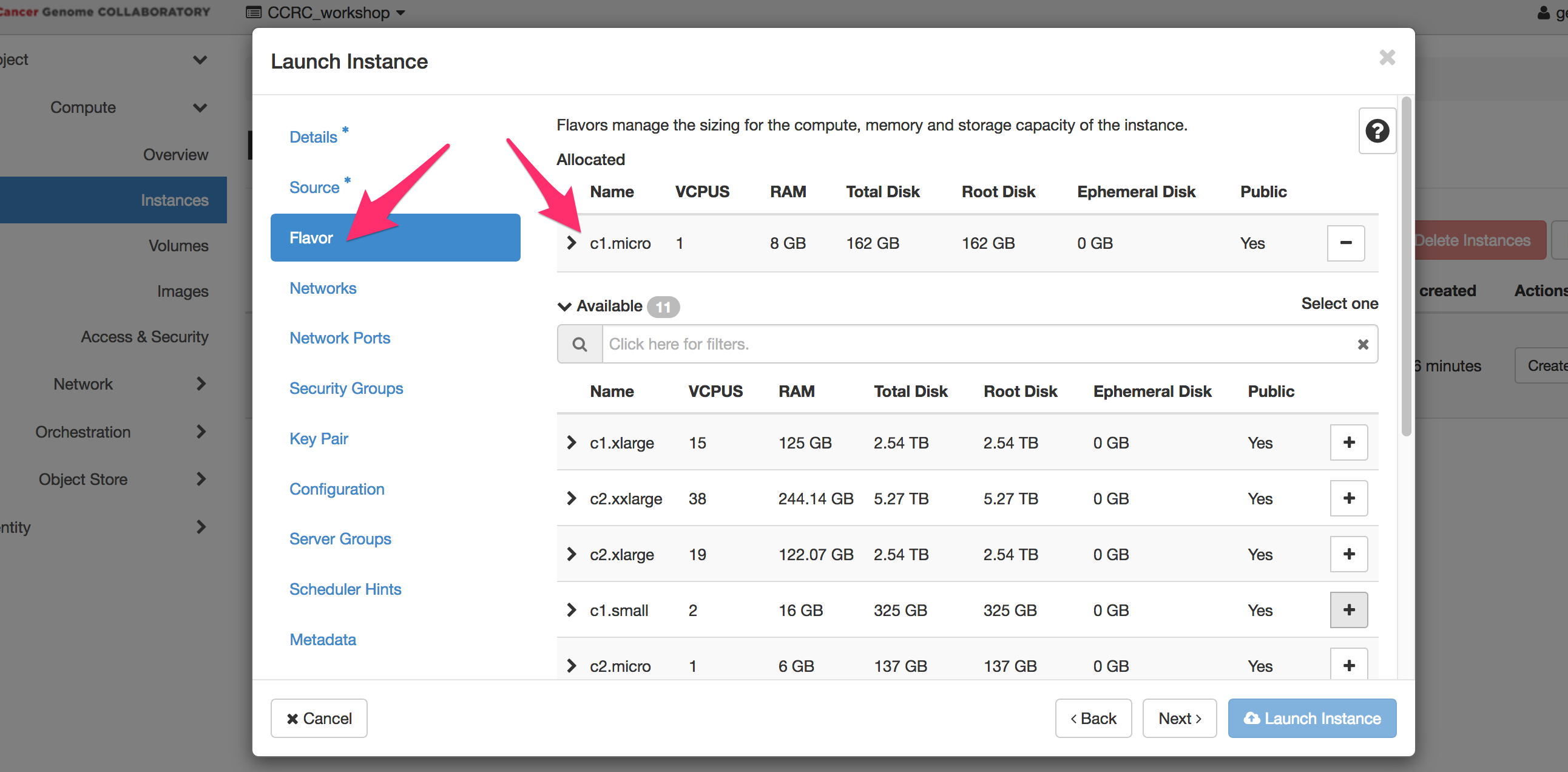
** Flavor: c1.micro **
** Network: CCRC_workshop_net (it should be already selected) **

** Network Ports: nothing to change here **
** Security group: default **

** Key Pair: your keypair’s name (it should be already selected) **
Leave the other tabs as they are and launch the instance by hitting the “Launch” button.

It will take a few minutes for the instance to start.
To view your instances, in the left hand menu, click on “Instances”.

Associate a Floating/Public IP Address
By default, the VM will have a private IP allocated that is only reachable from inside your virtual network (other VMs connected to the same network).
It is recommended to associate a floating/public IP address only to a single VM and use that one as a “jumpserver”.
From the “Compute” menu, select “Instances.” Beside the name of your instance, select “Associate Floating IP.” and follow the screenshots below to associate a floating IP with your virtual machine.



SSH Into Your Instance
Mac/Linux Instructions
You will need to change the file permissions for your private SSH key.
chmod 400 path_to_private_key
Where path_to_private_key is the path to where you have saved your key after you created it in the first steps.
Then, to log in:
ssh -i path_to_private_key ubuntu@142.1.177.XXX
XXX is the last octet from the floating IP address you assigned to the instance.
Windows Instructions
To configure Putty, start Putty and do the following:
- Fill in the “Host name” field with 142.1.177.XXX.
XXX is the last octet from the floating IP address you assign to the instance.
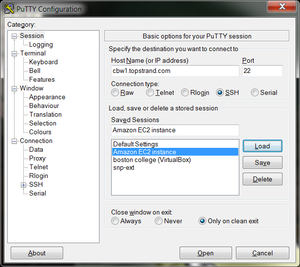
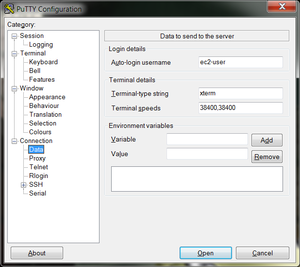
- In the left hand categories, under the Connection category choose Data. In the auto-login username field write ubuntu.
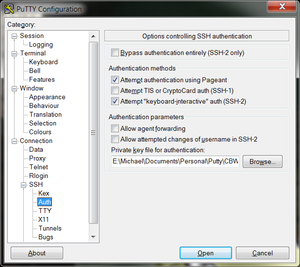
- In the left hand categories, in the Connection category next to SSH click on the +. Click on Auth. In the private-key file for authentication field, hit browse and find the private key that you converted previously from a PEM format to a PPK format.
- In the left hand categories, click on Session. In the Saved Sessions field write Collaboratory and click save.
Now that Putty is configured, all you have to do is start PuTTY and double-click on “Collaboratory” to login.
Customize Your Virtual Machine
You will first need to upgrade your package index and existing packages by running:
sudo apt-get update && sudo apt-get upgrade
Docker Installation
Run the following commands to install the Docker repository and required dependencies:
sudo apt-get install -y apt-transport-https ca-certificates unzip
curl -fsSL https://download.docker.com/linux/ubuntu/gpg | sudo apt-key add -
sudo add-apt-repository \
"deb [arch=amd64] https://download.docker.com/linux/ubuntu \
$(lsb_release -cs) \
stable"
sudo apt-get update
The following commands will install Docker and run hello-world to test the installation:
sudo apt-get -y install docker-ce
sudo service docker restart
sudo docker run hello-world
Run a Bioinformatics Tool in Docker
You will first need a data file. To get the file from the ftp server, we will use the wget command.
wget ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase3/data/NA12878/alignment/NA12878.chrom20.ILLUMINA.bwa.CEU.low_coverage.20121211.bam
You will need the Docker container with the “bamstats” tool. Please use the Docker pull command to retrieve the container.
sudo docker pull quay.io/briandoconnor/dockstore-tool-bamstats
Run the container, mounting the sample file inside.
sudo docker run -it -v `pwd`/NA12878.chrom20.ILLUMINA.bwa.CEU.low_coverage.20121211.bam:/NA12878.chrom20.ILLUMINA.bwa.CEU.low_coverage.20121211.bam -v /tmp:/home/ubuntu quay.io/briandoconnor/dockstore-tool-bamstats
In this command, -it means run interactively and -v maps a file or directory from the VM inside the Docker container. Whatever is created inside “/home/ubuntu” (inside the container) will be in the host “/tmp” directory. This will allow the files that are created inside the container to survive after the container is terminated.
The OS inside the Docker container is different than that of the host VM, and you can check this with:
cat /etc/lsb-release
Recall that you chose Ubuntu 16.04 for our VM, but the container is built based on a Ubuntu 14.04 image.
Inside the Docker container, execute the bamstats binary against the sample file.
cd && /usr/local/bin/bamstats 4 /NA12878.chrom20.ILLUMINA.bwa.CEU.low_coverage.20121211.bam
Exit the docker container by typing “exit”, and go to “/tmp” where the report was created:
exit
cd /tmp
unzip bamstats_report.zip
Start a simple Python http server that will allow you to see the bamstats report in the browser:
sudo python3 -m http.server 80
Visit the page to see the statistics for that sample BAM: http://142.1.177.XXX/bamstats_report.html
XXX is the last group of numbers from the floating IP address you assigned to the instance.
To stop the Python http server, press Ctrl+C.
Access Data in the Cloud
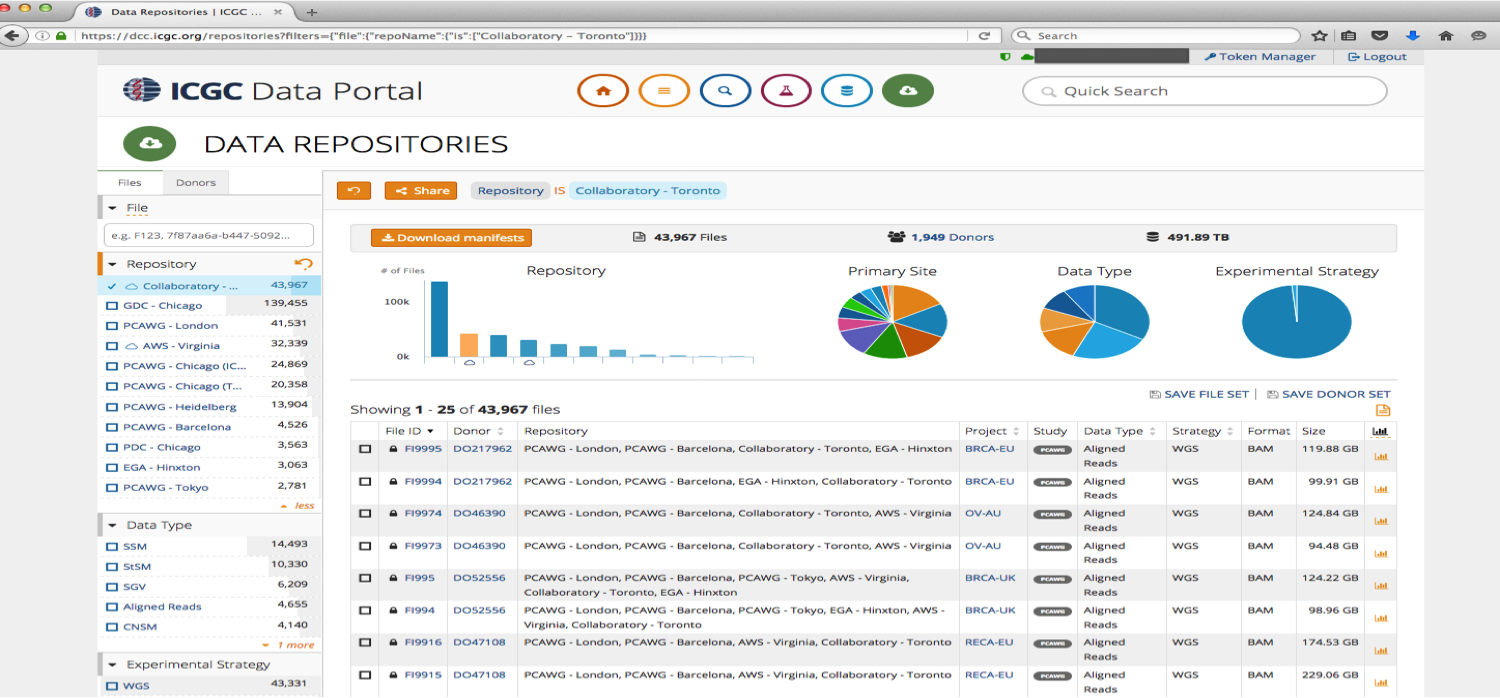
ICGC data (stored at GNOS sites, Collaboratory, AWS S3) and TCGA (stored at cgHub and GDC) are indexed at https://dcc.icgc.org/repositories.
Other data sets are usually available on-line and can be accessed over HTTP(s) protocol.
Data repositories have different access policies and download clients: http://docs.icgc.org/cloud/repositories/#download-client-operation_1
OICR created a unified client that can be used to download data from multiple repos http://docs.icgc.org/cloud/icgc-get/, but for this lab we will use the native “ICGC storage client” that can be used only in Collaboratory and AWS.
Collaboratory Data
In order to access ICGC data stored in the Cloud you need DACO approval, but for this workshop we uploaded a set of BAM files that are open access which can be used to test the functionality of the storage client. The same methodology can be used to download protected data, if a valid access token is used.
Below is the list of open-access BAM files and their object IDs stored in Collaboratory:
HCC1143.bam.bai dae332b8-107f-5e58-aefa-5c00de5d0bb3
HCC1143_BL.bam 561d9ddf-bbbc-5988-91c5-3c46d0d67ebe
HCC1143_BL.bam.bai 40c1b7a1-980e-57c5-9b82-538f1fdd5fc1
HCC1143_BL.RG1.bam caa34f59-f93f-53d3-aca0-03a18b8f7892
HCC1143_BL.RG2.bam 09a31178-48e9-5632-9085-a8a34c6144e3
HCC1143_BL.RG3.bam 08a65e40-4ff0-547d-89cf-b0a2859a5602
HCC1143_BL.RG4.bam ddd9d9e0-ac67-50b4-8a57-d8854372269c
HCC1143_BL.RG5.bam 3825dd85-cd0c-5121-9c05-0ce3e5883a08
HCC1143_BL.RG6.bam fd858cf7-e19b-566a-98cc-63516d607f2d
HCC1143_BL.RG7.bam 9ea3f173-2c6a-5dc8-974c-a0887f99c15f
HCC1143_BL.RG8.bam 75931e5b-ac08-57d6-b6d2-792e1609c588
HCC1143_BL.RG9.bam cff12c98-3ce7-5afa-8509-ba4a69880119
HCC1143_BL.RG10.bam 95ce9fda-b0aa-5e79-ae2b-74555031fee4
HCC1143_BL.RG11.bam 350d1d46-ddd1-5d67-bad0-94b29358cd2a
HCC1143_BL.RG12.bam 927dd39e-1199-55ac-b051-58b738c88bcb
HCC1143_BL.RG13.bam 147356e1-80cb-598a-ad0b-86007b92cb5e
HCC1143_BL.RG14.bam b5e1eb25-c297-53f9-b04f-0261b7c2a1f7
HCC1143_BL.RG15.bam 9717921b-67f3-5914-8c66-db3ef1ad6d61
HCC1143_BL.RG16.bam 891f33b1-b355-525e-a4e4-87a0397f5be4
Storage Client
In addition to a token scoped for the right environment (AWS or Collaboratory), you need a storage client in order to download ICGC data.
The storage client is provided as a tarball or packaged in a Docker container (http://docs.icgc.org/cloud/guide/).
Advanced functionality provided by the storage client:
- Fast parallel download of remote objects with option to resume an interrupted download session
- Single file download or manifest based download
- Generate a pre-signed temporary URL (24 hours) for a file that can be used to download with WGET or cURL
Create a Configuration File
By default, the storage client only uses a single cpu for its operation, so it’s important to tune its configuration to your local environment if you run it on a virtual machine with mutiple CPUs.
First, determine how many cores and how much memory you have available.
cat /proc/cpuinfo | grep -c processor
free -g
Considering that usually there is no analysys going until the data is retrieved, it is safe to allocate 7 cores and 49 GB of RAM (7 GB per thread) on a VM with 8 cores and 55 GB of RAM, leaving one CPU and 6 GB of RAM for the system. Create a text file /home/ubuntu/application.properties that contains the access token and the number of cores and memory per core as below. Use cat to view the file contents.
cat /home/ubuntu/application.properties
#accessToken=XXX ## you would have to uncomment this line and add a valid ICGC token in order to access protected-data. For this lab's purposes though, the client will download the files listed above even with the accessToken line commented out or missing
transport.parallel=7 ## use 7 paralled threads
transport.memory=7 ## use up to 7 GB of memory per thread to cache the downloaded data
How to Download Data using the storage client
Pull the Docker container containing the storage client.
sudo docker pull icgc/icgc-storage-client
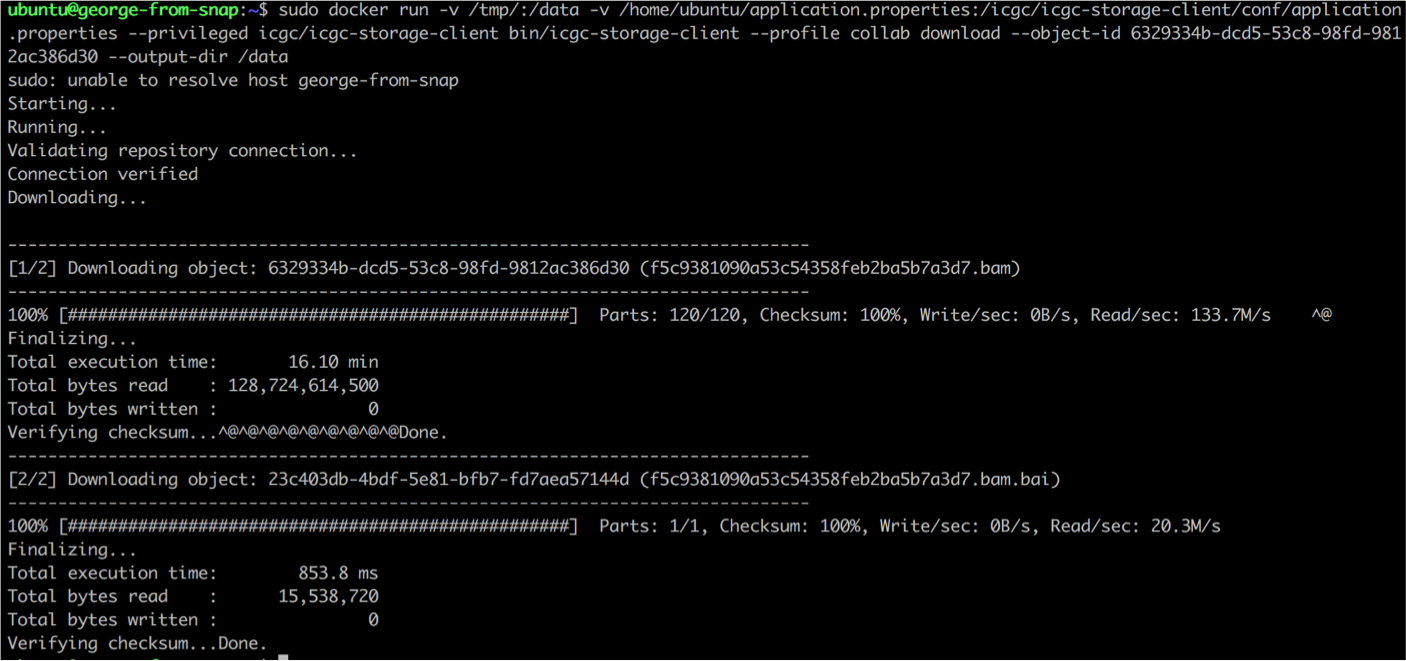
Choose an object_id from the list above containing open-data files, and initiate the download, mounting the application.properties file as well as the destination directory for the download (-v /tmp/:/data), so it survives the termination of the Docker container. Replace “OBJECT_ID” with one valid UUID from the list provided above.
sudo docker run -v /tmp/:/data -v /home/ubuntu/application.properties:/icgc/icgc-storage-client/conf/application.properties --privileged icgc/icgc-storage-client bin/icgc-storage-client --profile collab download --object-id OBJECT_ID --output-dir /data
As the files uploaded for this lab’s purposes are open access cell line BAM files, their size is smaller (~ 5 GB) so the download should complete in a couple of minutes.
Note: It takes around 16 min for a 100 GB file to be downloaded using a VM with 8 cores and 56 GB of RAM, and an additional 5-6 min is needed by the storage client to perform an automated checksum to verify downloaded data integrity.
The download time depends on the disk speed which is shared with other VMs running on the same physical server, as well as other shared resources (network load, storage cluster).
For a workflow that runs a few days, the 30 min needed to retrieve the data represent only a small fraction.
Important Notes
ICGC data stored in AWS S3 is only available from EC2 instances within the “us-east-1” EC2 region of AWS.
ICGC data stored in Cancer Genome Collaboratory is only available from VMs inside Collaboratory.
It is the responsibility of the users to protect access to the EC2 or Collaboratory VMs, as well as the restricted data they have access to.
Do not snapshot a VM that contains confidential tokens or protected data if the snapshot is intended to be shared with other cloud users. Also, keep in mind that other members of the same project as you could start instances from your snapshot and by doing so, have access to the data or tokens you left inside.
Congratulations, you have completed lab 1.
Note: Leave your VM running as it will be used in the second lab.