High-Throughput Biology - From Sequence to Networks Module 15 Lab Answers
This work is licensed under a Creative Commons Attribution-ShareAlike 3.0 Unported License. This means that you are able to copy, share and modify the work, as long as the result is distributed under the same license.
Module 15 Practical Lab: Reactome
By Robin Haw
CSHL-CBW Lab Module 15 Answers
Example 1
- EGFR, FGFR, SCF-KIT, PDGF, ERBB2, ERBB4, PI3K/AKT signaling, etc;
Example 2
- The overall sub-network consists of 303 nodes and 805 edges. The largest component of the subnetwork consists of 282 nodes and 723 edges, with the remainder of nodes and edges distributed amongst 5 other small subnetworks and interactions.
- Couple of ways to answer this. The driver mutations are probably the frequently mutated gene in the samples. The node size is proportional to the number of samples where the gene is mutated. Method 1- Look for the largest nodes in the diagram. Method 2 – Click Node Table and sort by “sampleNumber”. The largest node is TP53, ie. mutations in the TP53 gene are highly prevalent, occurring in 100 samples. Other driver mutations include EGFR (95) and PTEN (93). Additional mutations of interest include NF1, PIK3R1, PIK3CA, PIK3R1, RYR2, RB1.
- Search for “TP53 PEG3” in search bar in top right of Cytoscape tool. Annotated Functional Interaction based upon data from the TRED database. This targeted interaction describes an interaction between TP53 (regulator) and PEG3 (target). An immunoprecipitation experiment demonstrates the interaction, and the supporting evidence has been published in the paper with a PubMed ID: 11679586.
- Search for “TAF1 TAF7L” in search bar in top right of Cytoscape tool. Predicted Functional Interaction based upon data (2/9 sources are true) from a mouse interaction database and GO (GO BP sharing). FI score: 0.53
- 15 modules, with 9 modules of 10 ≥ genes.
- 20 modules, depending on the results of the enrichment analysis. Some pathways gene sets at the cutoff threshold may come or go but those highly significant gene sets are always there.
- 1: RTK signalling, 2: ECM and Integrin signalling, and 3: TP53 signaling and Cell Cycle.
Example 3
- The overall sub-network consists of 250 nodes and 517 edges. The largest component of the subnetwork consists of 212 nodes and 448 edges, with the remainder of nodes and edges distributed amongst 11 other small subnetworks and interactions.
- The largest node is TP53, ie. mutations in the TP53 gene are highly prevalent, occurring in at least 96% of HGS-OvCa samples.
- After clustering, there are 25 modules with 11 modules of 10 ≥ genes.
- 17 modules, depending on the results of the enrichment analysis. Some pathways gene sets at the cutoff threshold may come or go but those highly significant gene sets are always there.
- 0: ECM and Integrin signalling, 2: Calcium signalling-Adreneric Signaling-Cardiac Muscle Contraction, 3: Cell Cycle – Meiotic and Mitotic.
- Yes, ECM organization and Cell adhesion.
- Nuclear components - Nucleoplasm, nuclear membrane, nuclear pore, chromatin, etc.
- Modules 4, 5, 6 will be highlighted. Navigate through hierarchy. Neoplasm > Neoplasm_by_Site > Breast Neoplasm > Maligant_Breast_Neoplasm > Breast Carcinoma > Stage_IV_Breast_Cancer. Go back to the Network Module Browser. Genes in the modules that have ‘Stage IV Breast Cancer’ annotations will be yellow-highlighted: BRCA1, NRG1, TP53, INSR, EGFR.
- 3 modules: 0, 2, 8
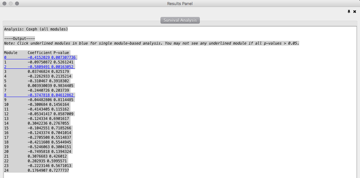
- The ReactomeFIViz app splits samples into two groups: samples having genes mutated in a module (red line), and samples having no genes mutated in the module (green line). The plugin uses the log-rank test to compare the two survival curves, and estimates p-values. In Modules 0 (KM: p= 0.00693), 2 (KM: p= 0.00141) and 8 (KM: p= 0.0451), patient with genes mutated (green line) have a better prognosis than patients with no gene mutations (red line). Module 2 is most statistically significant modules from the CoxPH and KM analysis.
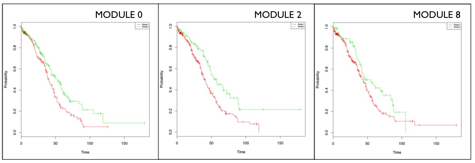
- In Module 2, the Calcium signaling, Chemical Synapse/Neurotransmission and Muscle Contraction annotations reflect a shared set of genes. These genes represent voltage-gated ion channels, which are a group of transmembrane ion channels that activated by changes in electrical potential difference. Even though ion channels are especially critical in neurons and muscle tissue, they are common in many types of cells, controlling the influx and outflux of ions. There are a number of genetic disorders, which disrupt normal functioning of ion channels. Calcium homeostasis is essential for cell migration, and tumor metastasis in particular. It may be that mutations in Module 2 genes disrupt calcium homeostasis, thereby impairing the tumour’s ability to metastasize, and extending patient’s overall survival.