High-Throughput Biology - From Sequence to Networks Pathways Integrated Assignment Answers
This work is licensed under a Creative Commons Attribution-ShareAlike 3.0 Unported License. This means that you are able to copy, share and modify the work, as long as the result is distributed under the same license.
Pathway and Network Analysis Integrated Assignment: Answers to questions and Screenshots
By Veronique Voisin
The results are stored in this folder.
g:Profiler
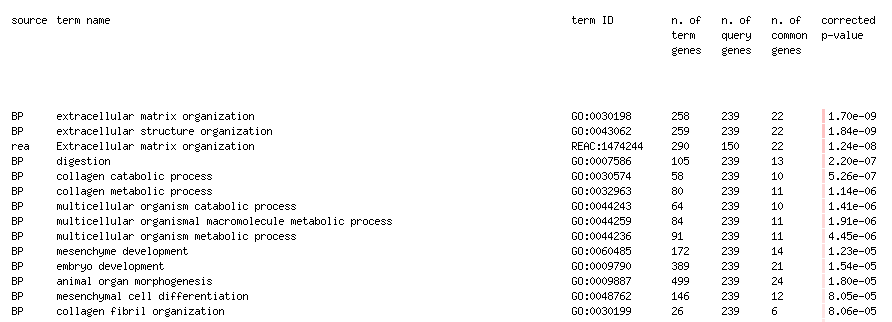
Question: What is the most significant GO:term? What is the p-value for this GO:term
Answer: extracellular matrix organization: p-value of 1.70e-09
Question: Is this p-value already corrected for multiple testing? What type of correction is used for your current analysis?
Answer: yes, it is already corrected for multiple hypothesis testing. I set the Significance threshold box to “Benjaminin-Hochberg FDR”.
g:Profiler png output
Re-run the analysis with User p-value threshold set to 0.0001.
Question: What has been changed?
Answer: Only the gene-set with pvalue equal or less than 0.0001 are displayed. The list is reduced compared to the results obtained with the default settings.
Ordered query:
Question: Do you seen any changes in the output in comparison to the analysis of the unordered gene list (PART 2)
Answer Although some terms are similar, their pvalues changed as well as the number of term genes used to calculate the pvalue.
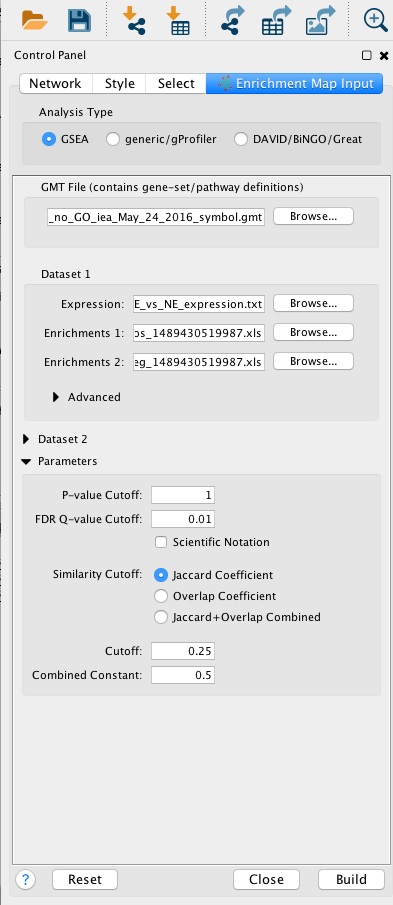
EnrichmentMap input panel
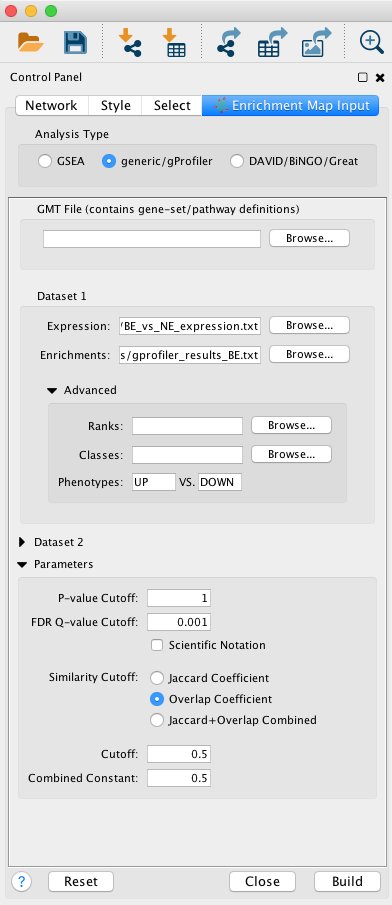
Question What can you conclude about these networks?
Answer The pathways are relevant to the biological model under study. The changes are related to the transformation of the epithelial cells into mesenchymal ones.
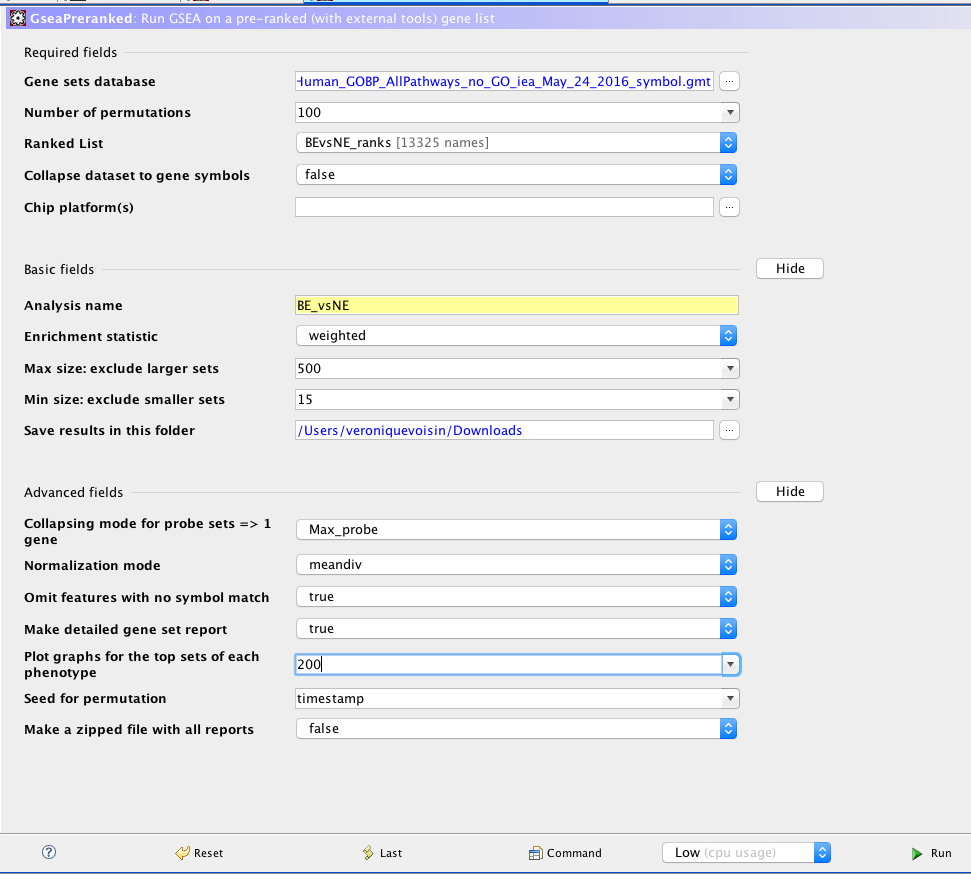
GSEA
GSEA input panel
GSEA EnrichmentMap input panel
DATASET2
PART1 REACTOME FI
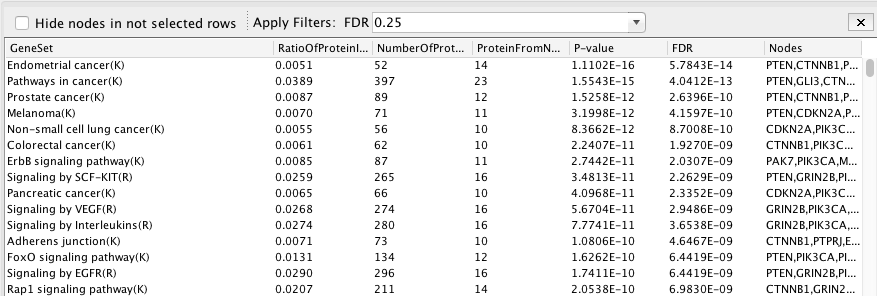
Pathway enrichment on the whole network.
Question What is the pathway with the lowest (best) FDR?
Answer The pathway with the lowest FDR is Endometrial cancer (K) with an FDR equal to 5.78w-16.
Cluster the network and perform pathway enrichment on the network.
Question How many clusters did the analysis retrieve?
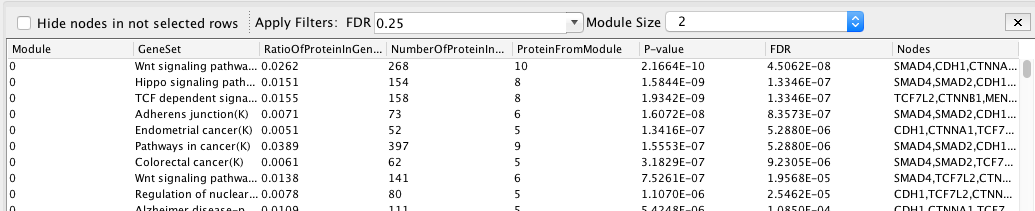
Answer The analysis retrieved 8 clusters named module 0 to module 7.
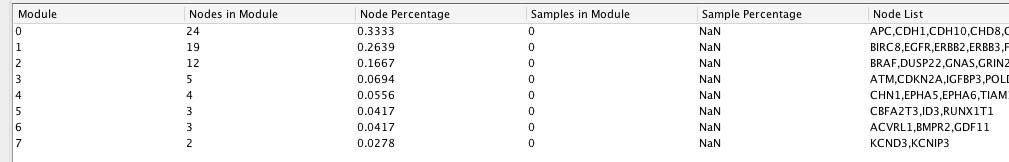
Question What is the FDR value of the most significant pathway of module 0?
Answer Wnt signaling pathway with FDR equal to 4.5e-08
PART2 GeneMANIA
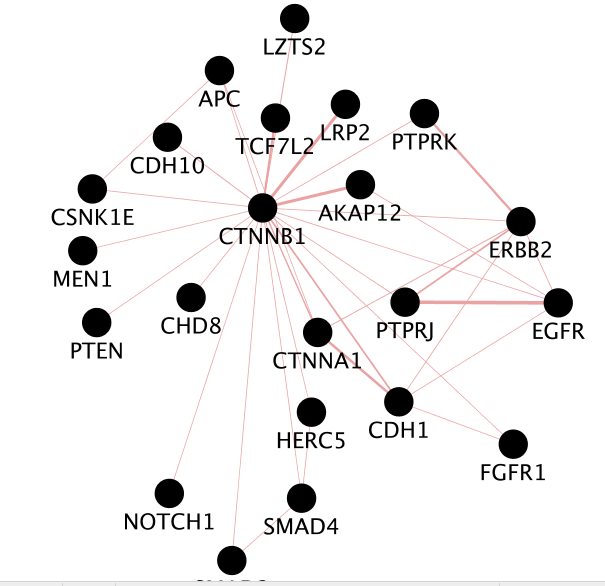
Question What is the number of nodes in the CTNBB1 network.
Answer There are 22 nodes.