So how do I process my data?
Analysis workflows
As the modules mentioned, there are multiple workflows that have been developed to help you process your data. The Broad Institute has developed a set of best practices when doing certain analysis as part of it’s Genome Analysis Toolkit. Flowcharts for their pipelines are available at their website, and cover:
- Data pre-processing
- Detecting somatic SNVs/Indels
- Detecting germline SNVs/Indels
- Detecting somatic CNVs
In addition to the workflows determined by the Broad institute, a few other workflows can be found here. These are meant :
-
RNA-Sequencing pipeline
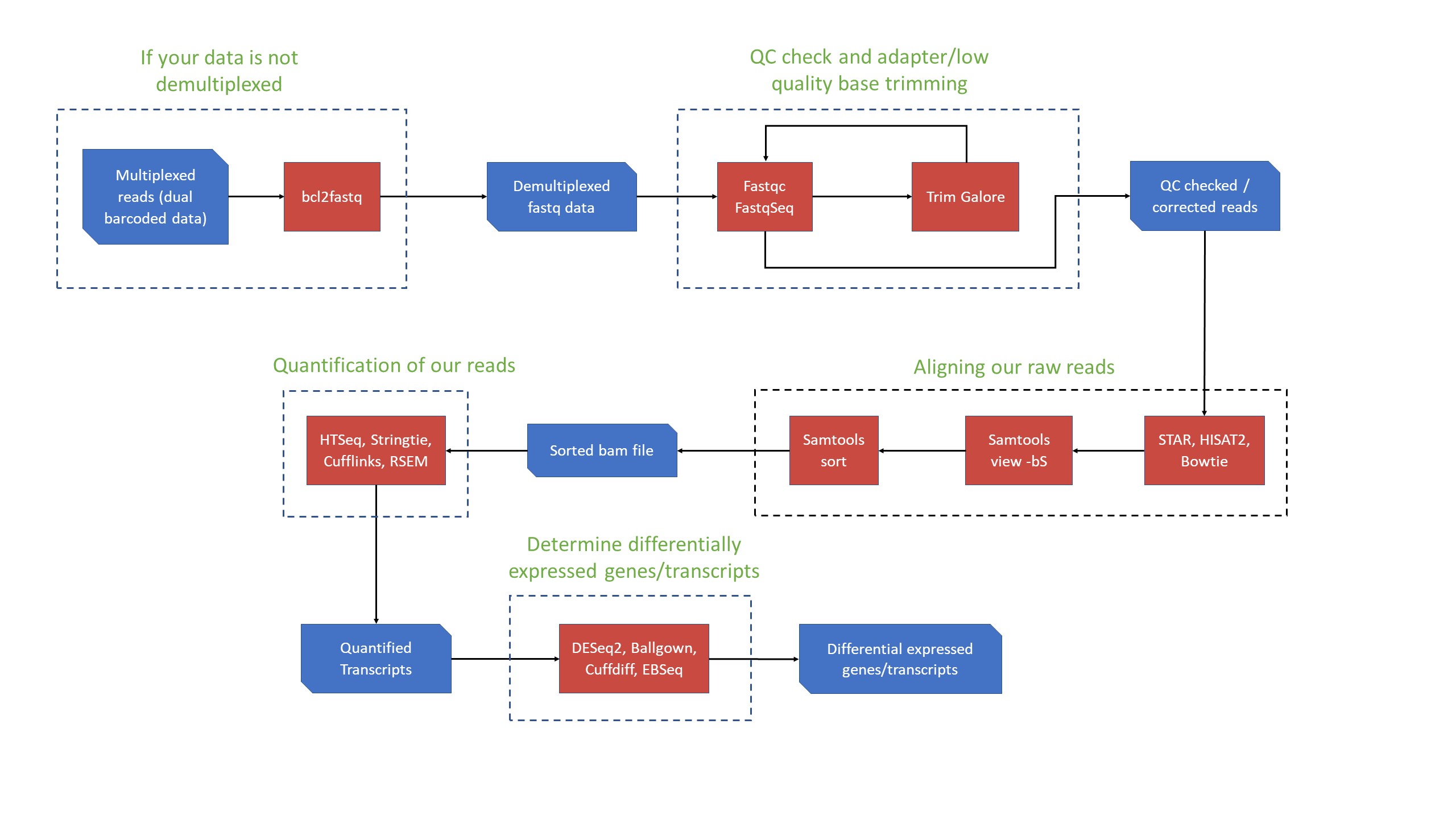
-
SNV Detection Workflow
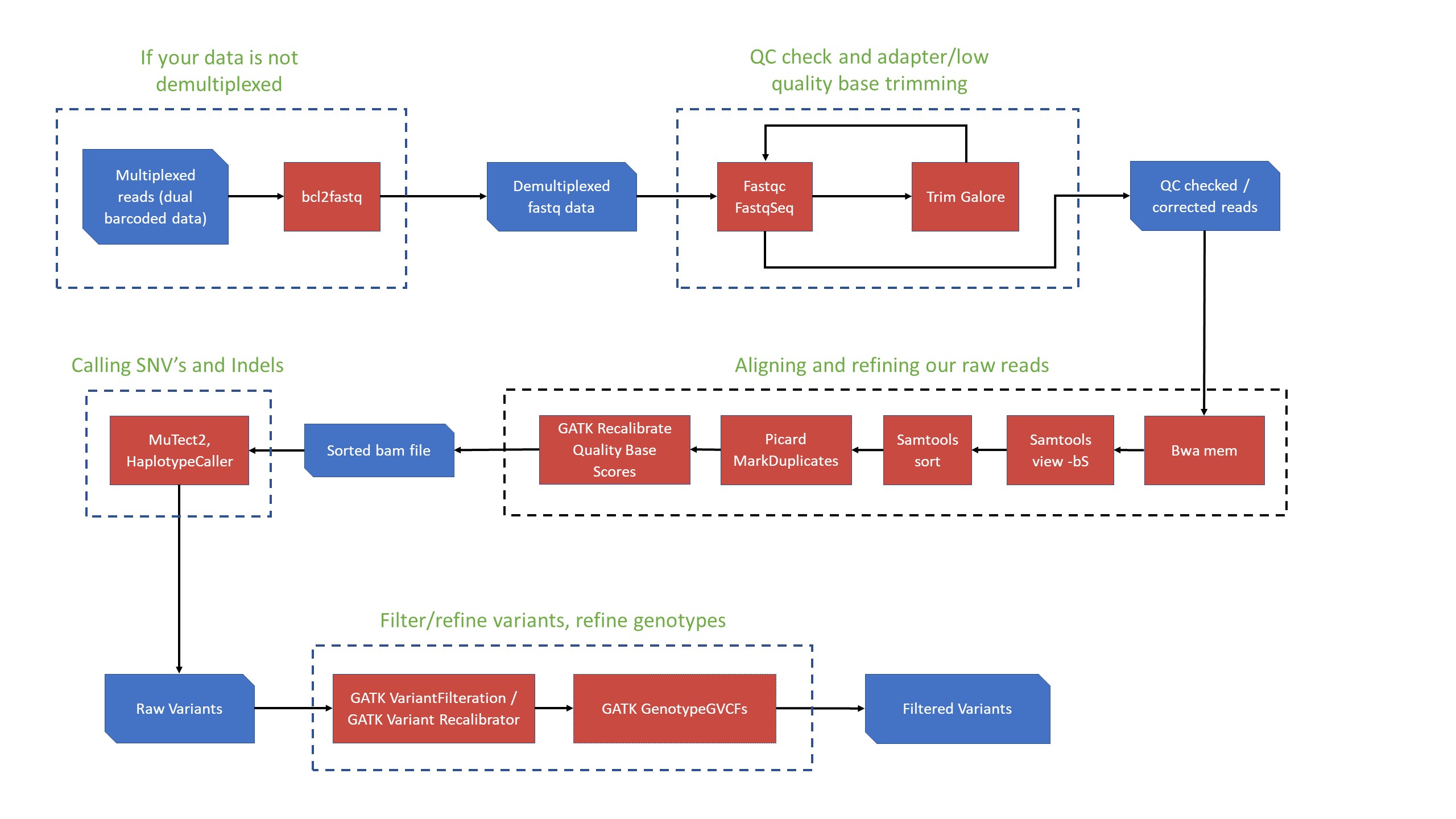
-
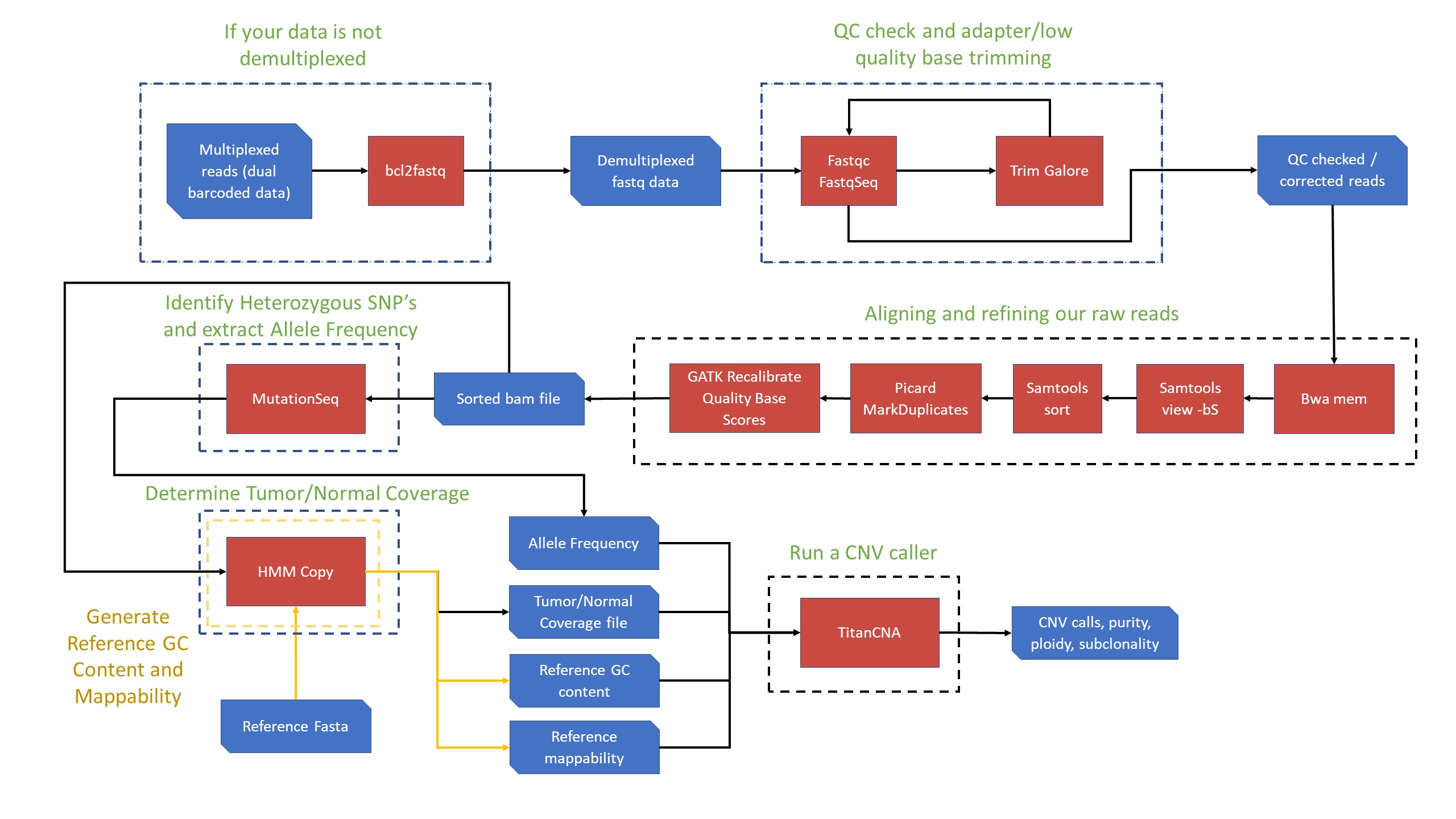
CNV Detection Workflow
File formats
- GenBank Flat File: file format for genbank intended to be human readable
- Fasta: sequence(s)
- Fastq: sequence(s) with qualities
- Sam/Bam: aligned reads either human readable or not
- VCF: variant file
For definitions of file formats, check here.
Databases
- many, many resources including literature (PubMed and PMC), health (dbGaP and ClinVar), genomes (WG adn RefSeq), proteins (Seq and 3D structures), and chemicals (PubChem and BioSystems)
- includes Entrez gene, the best way to explore gene information (according to BFFO)
- provides tools for visualizing, querying and downloading the data released quarterly by the consortium’s member projects.
- protected and non-protected data from 90 global cancer projects
- includes tumor/normal pairs with genome, transcriptome, methylome, and clinical data
- includes TCGA data
COSMIC: Catalogue of Somatic Mutations in Cancer
European Genome-Phenome Archive (EGA)
GDC: Genomic Data Commons Data Portal
Variation databases
Gene fusion databases
Genome browsers
Used to explore genomes. Can load “tracks” of extra info. Most common UCSC then EnsEMBL then NCBI Map Viewer. Many others exist.
Tools
Tools for CNVs
CRMAv2: http://www.aroma-project.org/vignettes/CRMAv2
DNACopy: http://www.bioconductor.org/packages/release/bioc/html/DNAcopy.html
HMM-Dosage: http://compbio.bccrc.ca/software/hmm-dosage/
PICNIC: http://www.sanger.ac.uk/genetics/CGP/Software/PICNIC/
OncoSNP: https://sites.google.com/site/oncosnp/
HMMCopy: http://compbio.bccrc.ca/software/hmmcopy/
Apolloh: http://compbio.bccrc.ca/software/apolloh/
Control-FreeC: http://bioinfo-out.curie.fr/projects/freec/
ASCAT: https://www.crick.ac.uk/peter-van-loo/software/ASCAT
ABSOLUTE: http://software.broadinstitute.org/cancer/software/genepattern/modules/docs/ABSOLUTE/1
TITAN: http://compbio.bccrc.ca/software/titan/
Tools for SNVs
GATK: http://www.broadinstitute.org/gsa/wiki/index.php/The_Genome_Analysis_Toolkit
SamTools: http://samtools.sourceforge.net/
MuTect: http://www.broadinstitute.org/software/cprg/?q=node/34
MuTect2: https://software.broadinstitute.org/gatk/documentation/tooldocs/3.8-0/org_broadinstitute_gatk_tools_walkers_cancer_m2_MuTect2.php
Strelka (from Illumina): https://sites.google.com/site/strelkasomaticvariantcaller/
SomaticSniper: http://gmt.genome.wustl.edu/packages/somatic-sniper/
VarScan: http://varscan.sourceforge.net/
Tools for Annotation
ANNOVAR: http://www.openbioinformatics.org/annovar/
SNPEFF: http://snpeff.sourceforge.net/
Pathway and Network Analysis
Command-line tricks
Pipe output for column view and scrolling
your command that would output to the screen in messy columns | column -t | less -S
column -t makes your tab delimited output into nice columns
less -S makes content in less left/right scrollable instead of wrapping it to the next line